
- Cytoscape rdf how to#
- Cytoscape rdf install#
- Cytoscape rdf software#
- Cytoscape rdf code#
- Cytoscape rdf download#
Posted in Uncategorized | Comments Closed Tags: cytoscape, semantic web, sparql, visualization
Cytoscape rdf code#
What’s even better is that the source code is available on github.Īlso, if you have a chance, come see my poster at Vizbi 2015 in Boston.
Cytoscape rdf download#
Or download it on from the app store website.
Cytoscape rdf install#
Just install Cytoscape 3.0, start it, and go to menu->Apps->App Manager and search for “General SPARQL”. The easiest way to get the app is simply from the Cytoscape App manager. The app works with privately managed triple stores just as well. But as great as these resources are, public triple stores can sometimes be overloaded. The initial set is designed to work with public bioinformatics SPARQL endpoints provided by the EBI and Bio2RDF. When you first install the app, it comes pre-configured with a basic set of SPARQL queries, although it’s possible to provide your own set. When you click on them, a query is sent off in the background, and the result is mapped to your network according to certain rules. Each item in the search menu, and each item in the context (right-click) menu, is backed by one SPARQL query. In the background, the General SPARQL app maintains a list of SPARQL queries. Watch this screencast and it will start to make sense: And you can continue this process, jumping from one entity to the next.
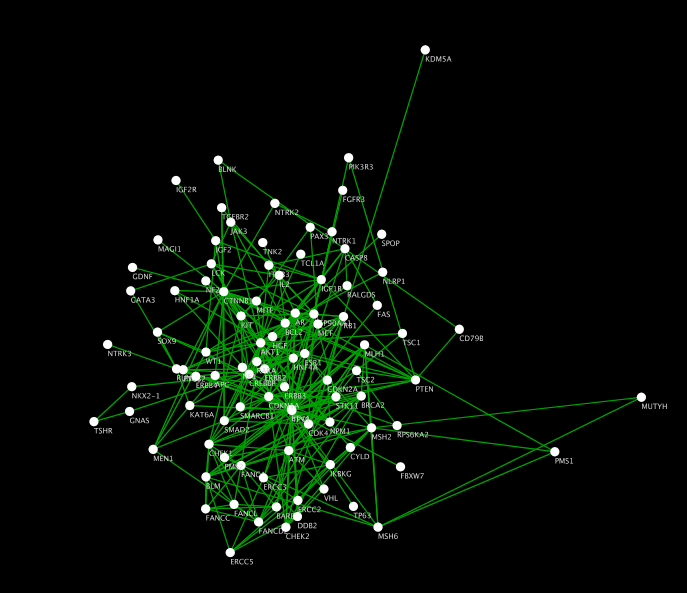
Or the gene that encodes for your protein. Or all the reactions that your protein is part of. For example, all the pathways that are related to your protein. You can then right-click on this node to pull in related entities. The app places a single node in your network. Edges can represent any type of relation between those entities.įor example, you can start by searching for a protein of interest. Nodes can represent various biological entities, such as: a pathway, a protein, a reaction, or a compound. Nodes and edges can be added to a network in a piecemeal fashion. The app lets you build a network step by step. The General SPARQL app is one of the new ways to present triple data. We shouldn’t expect scientists to write SPARQL queries anymore than we expect them to carry adjustable pliers to a restroom visit.

Cytoscape rdf software#
The problem is that SPARQL is supposed to be only a piece of plumbing at the bottom of a software stack. The problem isn’t that SPARQL is hard to use per se (it’s really rather plain and sensible). Integrating data is only half of the problem, we also have to present that data.
Cytoscape rdf how to#
Triple stores are great for data integration, but you still have to figure out how to put that data in the hands of scientists. But how do we put that data in the hands of scientists?Īt General Bioinformatics we put data in triple stores, and use SPARQL to query that data. We can now easily solve the problem of bioinformatics data integration. The R kernel for Jupyter is available here, and the installation instructions are on the same page. What follows are some notes I took during that process, in the hope that they are useful for anybody else trying to do the same thing.įirst you need to have R and Jupyter installed, but I’m assuming you already got that far. It took me a while to get that working for the R scripting language. Jupyter consists of multiple ‘kernels’, to get support for a different language you have to install that language, and then install the Jupyter kernel for it. In principle you can install support for a wide range of scripting languages, but in practice it may be a little difficult to set up. But since they added support for other languages besides python, they had to rename. Jupyter started its life as IPython, or “interactive python”. A Jupyter notebook can contain the analysis, the results, and the documentation that explains the results together in a single file, making it at once understandable and reproducible. Jupyter notebooks are a great way to do an analysis, and report the results at the same time.
Scripts are a great way to make reproducible workflows, but they are too technical for many situations where you have to report to scientists. Teach a scientist to script and they won’t have any more time to do analysis for the rest of their lifetime. Give a scientist a script and they will analyse data for a day.


 0 kommentar(er)
0 kommentar(er)
